Microarray Analysis Of Products of Conception (POC)
 Microarray-based Comparative Genomic Hybridization (aCGH) is a new genetic test that can detect
both unbalanced genomic alterations usually identified by chromosome analysis (karyotyping) and
unbalanced genomic alterations that cannot be identified by karyotyping (including microdeletions
and microduplications). Microarray can be performed directly on DNA from POC tissue without cell
culturing. CGH+SNP microarrays can simultaneously detect copy number changes as well as copy
neutral aberrations, such as absence of heterozygosity (AOH) and uniparental isodisomy (UPD).
Cases of triploidy and complete molar pregnancies that are completely homozygous and of androgenetic
origin can be readily diagnosed with CGH+SNP.
Microarray-based Comparative Genomic Hybridization (aCGH) is a new genetic test that can detect
both unbalanced genomic alterations usually identified by chromosome analysis (karyotyping) and
unbalanced genomic alterations that cannot be identified by karyotyping (including microdeletions
and microduplications). Microarray can be performed directly on DNA from POC tissue without cell
culturing. CGH+SNP microarrays can simultaneously detect copy number changes as well as copy
neutral aberrations, such as absence of heterozygosity (AOH) and uniparental isodisomy (UPD).
Cases of triploidy and complete molar pregnancies that are completely homozygous and of androgenetic
origin can be readily diagnosed with CGH+SNP.
We provide CGH+SNP and High Resolution X-chromosome microarray (X-HR) tests on samples from products of conception.
Samples For Testing
The microarray testing will be performed using DNA samples extracted directly from the fetal tissue or from cultured fetal cells. In order to identify triploids that are partial moles and androgenetic heterozygous complete moles arising by dispermy at least one parental sample such as decidua would be required.
Whole Genome CGH+SNP Analysis
Clinical Indications For CGH+SNP Analysis
- Recurrent miscarriages
- An abnormal fetal karyotype requiring further characterization
- Failure to establish growth in culture due to maceration, infection, or prior fixation
- Parental consanguinity to identify possible candidate loci for recessive disorder
- An abnormal fetal karyotype requiring further characterization
- Stillbirth/fetal loss
- Suspected partial or complete molar pregnany
- Fetal abnormalities with apparently balanced chromosome rearrangement or unidentified marker chromosome
- Suspected triploidy
Platform For CGH+SNP Analysis
We use Agilent’s SurePrint G3 CGH+SNP microarrays (4x180K ISCA design) platform. The 110,712 (CGH) oligo probes and 59,647 (SNP) probes with 25.3 kb overall median probe spacing are throughout the genome and with 5 kb in ISCA regions. This platform is designed based on UCSC hg19 (NCBI Build 37, Februry 2009).
High Resolution X-Chromosome Microarray Analyses (X-HR)
Clinical Indication X-HR Analysis
- Fetus with suspected X-linked genetic disorder
- Fetus with suspected ambiguous genitalia
Platform For X-HR
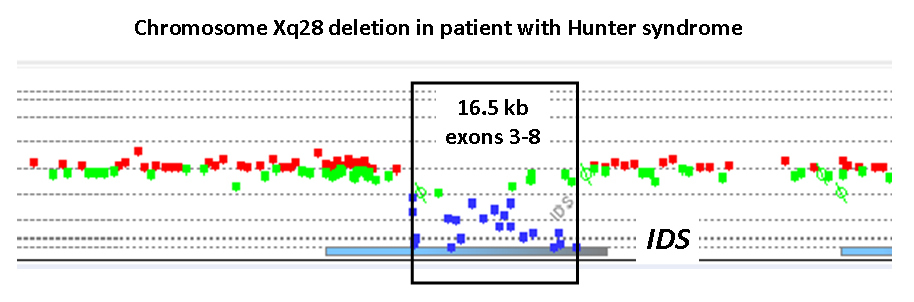 We use the Agilent 180K oligonucleotide array platform specifically designed and validated by Pittsburgh
Cytogenetics Laboratory for the X-chromosome disorders. The 180,000 oligonucleotides on the X-HR chip cover
the entire X-chromosome genome with high density probes in the regions containing genes involved in X-chromosome
disorders or associated with premature ovarian failure, as well as some autosomal genes involved disorders of
sexual differentiation. The maximum probe spacing is one probe for every 1 Kb throughout the X-chromosome
genome and one probe for every 0.3-0.5 Kb in the regions containing genes.
We use the Agilent 180K oligonucleotide array platform specifically designed and validated by Pittsburgh
Cytogenetics Laboratory for the X-chromosome disorders. The 180,000 oligonucleotides on the X-HR chip cover
the entire X-chromosome genome with high density probes in the regions containing genes involved in X-chromosome
disorders or associated with premature ovarian failure, as well as some autosomal genes involved disorders of
sexual differentiation. The maximum probe spacing is one probe for every 1 Kb throughout the X-chromosome
genome and one probe for every 0.3-0.5 Kb in the regions containing genes.
Advantages And Limitations For CGH+SNP Test
CGH+SNP can:
- simultaneously and rapidly evaluate thousands of loci for copy number imbalances
- further characterize the chromosome imbalances detected by karyotyping (e.g., maker chromosomes)
- detect absence of heterozygosity (AOH) and uniparental isodisomy (UPD)
- detect polyploidies such as complete mole or partial mole pregnancies
CGH+SNP cannot detect:
- balanced rearrangements (e.g., balanced translocation, inversion)
- base pair mutations
- gains or losses in the regions of the genome not covered by array
- low level mosaicism (<20%)
Microarray Analysis Of Products of Conception (POC)
Microarray-based Comparative Genomic Hybridization (aCGH) is a new genetic test that can detect both unbalanced genomic alterations usually identified by chromosome analysis (karyotyping) and unbalanced genomic alterations that cannot be identified by karyotyping (including microdeletions and microduplications). Microarray can be performed directly on DNA from POC tissue without cell culturing. CGH+SNP microarrays can simultaneously detect copy number changes as well as copy neutral aberrations, such as absence of heterozygosity (AOH) and uniparental isodisomy (UPD). Cases of triploidy and complete molar pregnancies that are completely homozygous and of androgenetic origin can be readily diagnosed with CGH+SNP.
We provide CGH+SNP and High Resolution X-chromosome microarray (X-HR) tests on samples from products of conception.
Samples For Testing
The microarray testing will be performed using DNA samples extracted directly from the fetal tissue or from cultured fetal cells. In order to identify triploids that are partial moles and androgenetic heterozygous complete moles arising by dispermy at least one parental sample such as decidua would be required.
Whole Genome CGH+SNP Analysis
Clinical Indications For CGH+SNP Analysis
- Recurrent miscarriages
- An abnormal fetal karyotype requiring further characterization
- Failure to establish growth in culture due to maceration, infection, or prior fixation
- Parental consanguinity to identify possible candidate loci for recessive disorder
- An abnormal fetal karyotype requiring further characterization
- Stillbirth/fetal loss
- Suspected partial or complete molar pregnany
- Fetal abnormalities with apparently balanced chromosome rearrangement or unidentified marker chromosome
- Suspected triploidy
Platform For CGH+SNP Analysis
We use Agilent’s SurePrint G3 CGH+SNP microarrays (4x180K ISCA design) platform. The 110,712 (CGH) oligo probes and 59,647 (SNP) probes with 25.3 kb overall median probe spacing are throughout the genome and with 5 kb in ISCA regions. This platform is designed based on UCSC hg19 (NCBI Build 37, Februry 2009).
High Resolution X-Chromosome Microarray Analyses (X-HR)
Clinical Indication X-HR Analysis
- Fetus with suspected X-linked genetic disorder
- Fetus with suspected ambiguous genitalia
Platform For X-HR
We use the Agilent 180K oligonucleotide array platform specifically designed and validated by Pittsburgh Cytogenetics Laboratory for the X-chromosome disorders. The 180,000 oligonucleotides on the X-HR chip cover the entire X-chromosome genome with high density probes in the regions containing genes involved in X-chromosome disorders or associated with premature ovarian failure, as well as some autosomal genes involved disorders of sexual differentiation. The maximum probe spacing is one probe for every 1 Kb throughout the X-chromosome genome and one probe for every 0.3-0.5 Kb in the regions containing genes.
Advantages And Limitations For CGH+SNP Test
CGH+SNP can:
- simultaneously and rapidly evaluate thousands of loci for copy number imbalances
- further characterize the chromosome imbalances detected by karyotyping (e.g., maker chromosomes)
- detect absence of heterozygosity (AOH) and uniparental isodisomy (UPD)
- detect polyploidies such as complete mole or partial mole pregnancies
CGH+SNP cannot detect:
- balanced rearrangements (e.g., balanced translocation, inversion)
- base pair mutations
- gains or losses in the regions of the genome not covered by array
- low level mosaicism (<20%)